THE MOLECULAR CIRCUITS OF THE LIVING BRAIN:
Development, behavior and cognition are at once microscale processes and macroscale interactions between a living organism and its environment. To understand the living brain and its pathologies we must similarly scale from molecular mechanisms to whole brain function, which has proven to be one of the greatest challenges faced in neurobiology. Meeting this challenge demands an approach that can reveal molecular events across the brain, without disrupting the very processes we wish to study. The most powerful technology available for noninvasive whole brain molecular imaging is MRI, with the potential for subsecond and near cellular resolution. It is my overarching goal to realize the potential of MRI to resolve the diverse molecular signaling of development, function, and degeneration across the whole mammalian brain in vivo. To this end, I have established a versatile molecular toolkit for MRI, including calcium sensors and genetic reporters. Here, I propose hypothesis driven projects that focus the application of my methods on two forms of neurobiological signaling that offer the greatest impact on human health: calcium imaging of neural activity and neuroimmune interactions in regeneration. The work of my lab will establish the field of molecular fMRI, bringing powerful new capabilities to research by bridging functional and mechanistic biology.
PROBING THE NEUROIMMUNE REGENERATIVE SIGNALING AXIS WITH MRI REPORTER GENES:
PROBING THE NEUROIMMUNE REGENERATIVE SIGNALING AXIS WITH MRI REPORTER GENES: Stem cells build and replenish the mammalian brain through developmental tissue patterning and through adult neuronal regeneration, both basally, and in response to injury. Neuroimmune cells, like microglia, have an established neogenic role in healthy brain development, but the same cells are highly susceptible to external factors, leading to pathologies. Immune challenges affect development, while regeneration can be inhibited by inflammation, leading to degenerative disorders. Neuroimmune signaling remains extremely difficult to study in vivo given the sensitivity of microglia to the invasive methods needed to image the brain. New technologies for noninvasive functional studies would greatly accelerate research on any major neurodegenerative or inflammatory disease.
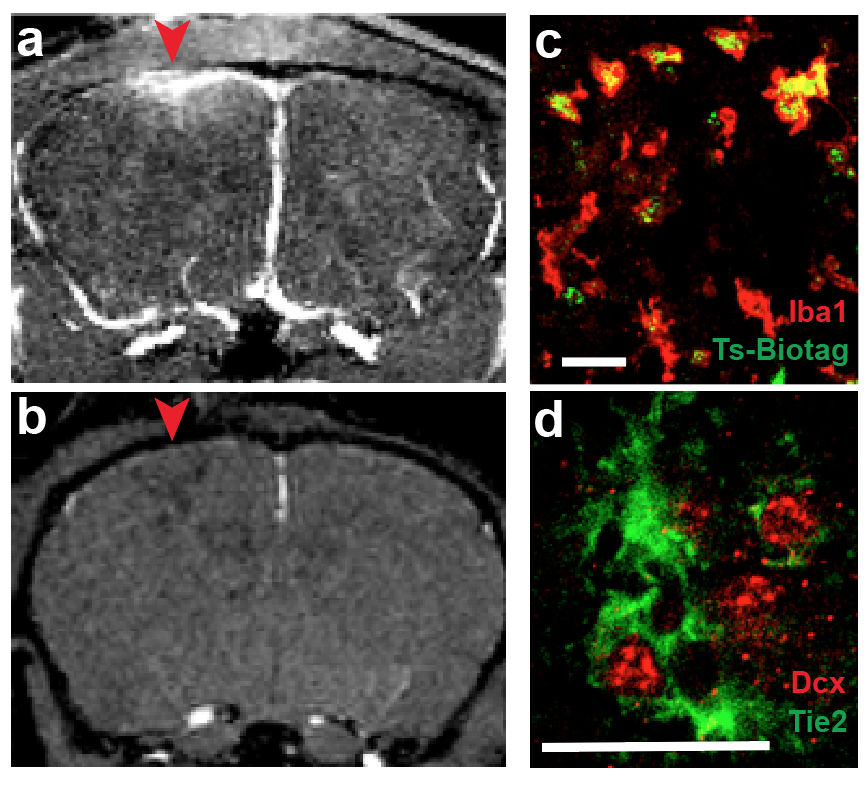
I have invented several genetic reporters for MRI and generated a transgenic mouse line that labels cell types responding to the neogenic cytokine Ang1. In preliminary studies, I have imaged activated microglia responding to a brain injury model (Fig. 1), an unprecedented functional study of a molecular circuit involved in neuroinflammation and adult neurogenesis. The proposed studies will further define capabilities of genetic imaging with MRI by using this established model to test a hypothesis that adult neogenic neuroimmune interactions recapitulate developmental signaling.The full diversity of cell types and signaling molecules in development and regeneration continues to be described, however the in vivo signaling dynamics of cytokines are virtually unknown, for lack of in vivo assays of function. Parsing neuroimmune signaling modes offers a new means to determine therapeutic targets for neurodegenerative disorders.